Variable Importance in RLT
RLT Package
2025-07-17
Variable-Importance.RmdIntroduction
Variable importance is a crucial aspect of random forest analysis that helps identify which variables are most influential in making predictions. The RLT package provides multiple approaches for calculating variable importance, including permutation-based methods and distributed assignment approaches.
Variable Importance Methods
1. Permutation-based Variable Importance
The permutation approach measures variable importance by randomly permuting the values of each variable and measuring the resulting change in prediction accuracy. This method requires a significant number of out-of-bag samples.
2. Distributed Assignment Variable Importance
This approach calculates all possible terminal nodes and their probabilities for a subject that could land in that terminal node. It sends observations down to both child nodes with weights proportional to the child node sizes, allowing calculation of variable importance for sampling without replacement with just one out-of-bag sample.
Regression Example
library(RLT)
#> RLT and Random Forests v4.2.6
#> pre-release at github.com/teazrq/RLT
# Generate sample data
set.seed(1)
n = 1000
p = 20
X = matrix(rnorm(n*p), n, p)
y = X[, 1] + X[, 2] + rnorm(n)
# Split data
trainn = 800
trainX = X[1:trainn, ]
trainY = y[1:trainn]
testX = X[(trainn+1):n, ]
testY = y[(trainn+1):n]
# Model parameters
ntrees = 100
mtry = 5
nmin = 5
nsplit = 10
ncores = 1
rule = "best"Permutation-based Variable Importance
# Fit RLT model with permutation importance
RLTfit <- RLT(trainX, trainY, model = "regression",
ntrees = ntrees, mtry = mtry, nmin = nmin,
split.gen = rule, nsplit = nsplit,
resample.prob = 0.8,
resample.replace = FALSE,
importance = "permute",
ncores = ncores,
verbose = TRUE)
#> Regression Random Forest ...
#> ---------- Parameters Summary ----------
#> (N, P) = (800, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.8 w/o replace
#> (Obs, Var) weights = (No, No)
#> importance = permute
#> reinforcement = No
#> ----------------------------------------
# Plot variable importance
par(mfrow=c(2,2))
par(mar = c(1, 2, 2, 2))
barplot(as.vector(RLTfit$VarImp), main = "RLT Permutation",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
Distributed Assignment Variable Importance
# Fit RLT model with distributed importance
RLTfit_dist <- RLT(trainX, trainY, model = "regression",
ntrees = ntrees, mtry = mtry, nmin = nmin,
split.gen = rule, nsplit = nsplit,
resample.prob = (trainn - 1)/trainn,
resample.replace = FALSE,
importance = "distribute",
ncores = ncores,
verbose = TRUE)
#> Regression Random Forest ...
#> ---------- Parameters Summary ----------
#> (N, P) = (800, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.99875 w/o replace
#> (Obs, Var) weights = (No, No)
#> importance = distribute
#> reinforcement = No
#> ----------------------------------------
# Plot distributed importance
par(mfrow=c(1,2))
par(mar = c(1, 2, 2, 2))
barplot(as.vector(RLTfit$VarImp), main = "RLT Permutation",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
barplot(as.vector(RLTfit_dist$VarImp), main = "RLT Distributed",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)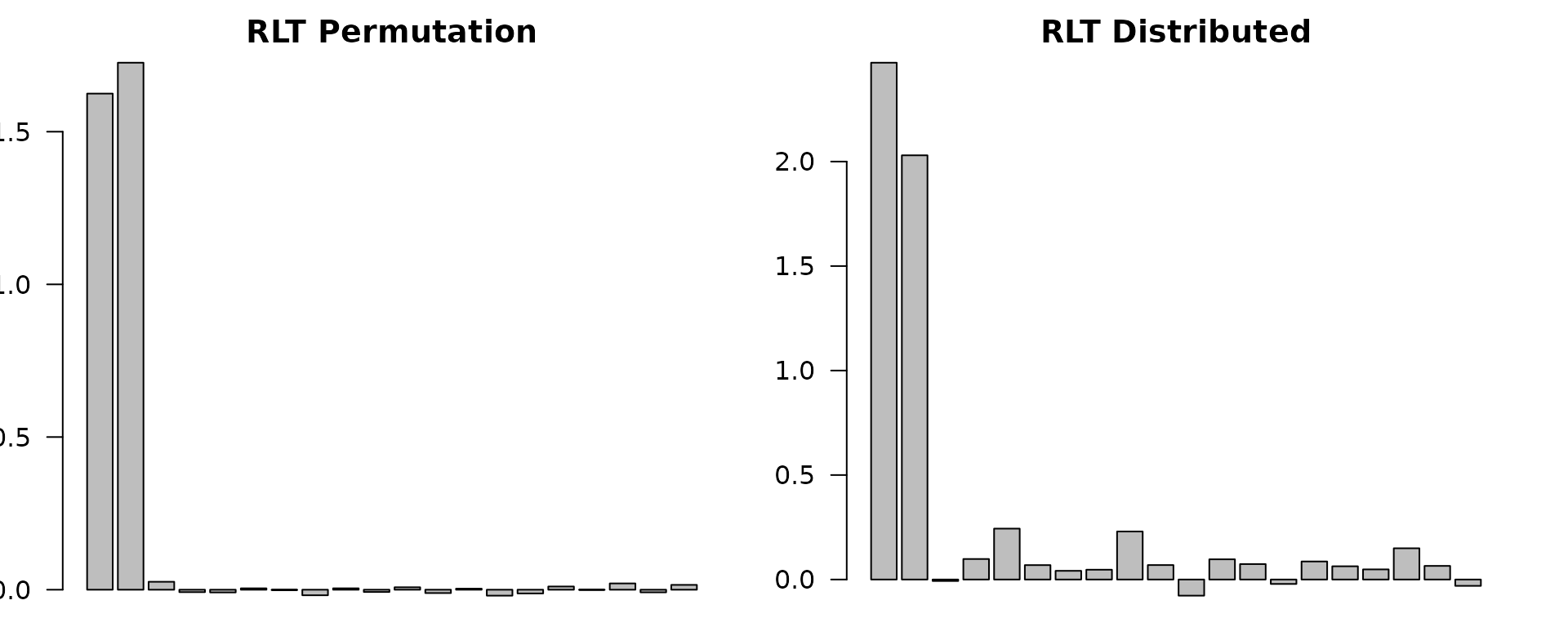
Classification Example
# Generate classification data
set.seed(1)
n = 1000
p = 20
X = matrix(rnorm(n*p), n, p)
y = as.factor(ifelse(X[, 1] + X[, 2] > 0, 1, 0))
# Split data
trainn = 800
trainX = X[1:trainn, ]
trainY = y[1:trainn]
testX = X[(trainn+1):n, ]
testY = y[(trainn+1):n]Classification Variable Importance
# Fit classification model with importance
RLTfit_cla <- RLT(trainX, trainY, model = "classification",
ntrees = ntrees, mtry = mtry, nmin = nmin,
split.gen = rule, nsplit = nsplit,
resample.prob = 0.8,
resample.replace = FALSE,
importance = "permute",
ncores = ncores,
verbose = TRUE)
#> Classification Random Forest ...
#> ---------- Parameters Summary ----------
#> (N, P) = (800, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.8 w/o replace
#> (Obs, Var) weights = (No, No)
#> importance = permute
#> reinforcement = No
#> ----------------------------------------
# Plot variable importance
par(mfrow=c(2,2))
par(mar = c(1, 2, 2, 2))
barplot(as.vector(RLTfit_cla$VarImp), main = "RLT Classification",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
# Distributed importance for classification
RLTfit_cla_dist <- RLT(trainX, trainY, model = "classification",
ntrees = ntrees, mtry = mtry, nmin = nmin,
split.gen = rule, nsplit = nsplit,
resample.prob = (trainn - 1)/trainn,
resample.replace = FALSE,
importance = "distribute",
ncores = ncores,
verbose = TRUE)
#> Classification Random Forest ...
#> ---------- Parameters Summary ----------
#> (N, P) = (800, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.99875 w/o replace
#> (Obs, Var) weights = (No, No)
#> importance = distribute
#> reinforcement = No
#> ----------------------------------------
barplot(as.vector(RLTfit_cla_dist$VarImp), main = "RLT Classification Distributed",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)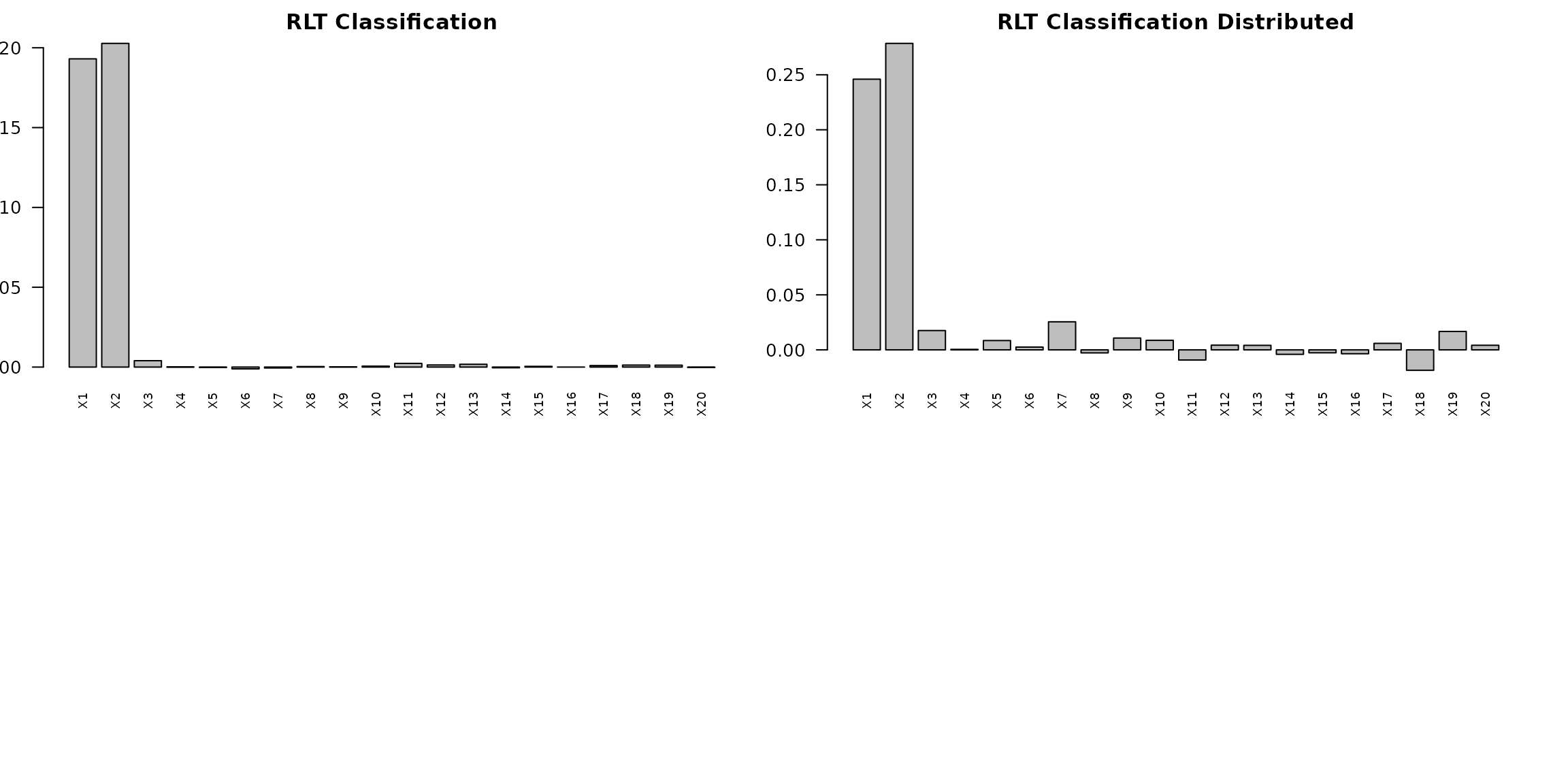
Survival Analysis Example
# Generate survival data
set.seed(1)
n = 600
p = 20
X = matrix(rnorm(n*p), n, p)
# Create survival times
xlink <- function(x) exp(x[, 1] + x[, 3]/2)
FT = rexp(n, rate = xlink(X))
CT = pmin(6, rexp(n, rate = 0.25))
Y = pmin(FT, CT)
Censor = as.numeric(FT <= CT)
# Split data
trainn = 500
trainX = X[1:trainn, ]
trainY = Y[1:trainn]
trainCensor = Censor[1:trainn]
testX = X[(trainn+1):n, ]
testY = Y[(trainn+1):n]
testCensor = Censor[(trainn+1):n]Survival Variable Importance
# Fit survival models with different splitting rules
RLTfit_logrank <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, ncores = ncores,
nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = 0.8,
importance = TRUE,
param.control = list(split.rule = "logrank", "alpha" = 0.2),
verbose = TRUE, resample.replace=FALSE)
#> Fitting Survival Forest...
#> ---------- Parameters Summary ----------
#> (N, P) = (500, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.8 w/o replace
#> (Obs, Var) weights = (No, No)
#> alpha = 0.2
#> importance = permute
#> reinforcement = No
#> ----------------------------------------
RLTfit_suplogrank <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, ncores = ncores,
nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = 0.8,
importance = TRUE,
param.control = list(split.rule = "suplogrank", "alpha" = 0),
verbose = TRUE, resample.replace=FALSE)
#> Fitting Survival Forest...
#> ---------- Parameters Summary ----------
#> (N, P) = (500, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.8 w/o replace
#> (Obs, Var) weights = (No, No)
#> importance = permute
#> reinforcement = No
#> ----------------------------------------
RLTfit_cg <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, ncores = ncores,
nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = 0.8,
importance = TRUE,
param.control = list(split.rule = "coxgrad", "alpha" = 0),
verbose = TRUE, resample.replace=FALSE)
#> Fitting Survival Forest...
#> ---------- Parameters Summary ----------
#> (N, P) = (500, 20)
#> # of trees = 100
#> (mtry, nmin) = (5, 5)
#> split generate = Best
#> sampling = 0.8 w/o replace
#> (Obs, Var) weights = (No, No)
#> importance = permute
#> reinforcement = No
#> ----------------------------------------
# Plot variable importance for different splitting rules
par(mfrow=c(2,2))
par(mar = c(1, 2, 2, 2))
barplot(as.vector(RLTfit_logrank$VarImp), main = "RLT Log-rank",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
barplot(as.vector(RLTfit_suplogrank$VarImp), main = "RLT Sup Log-rank",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
barplot(as.vector(RLTfit_cg$VarImp), main = "RLT Cox-grad",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
Comparison with Other Packages
# Load required packages for comparison
if (requireNamespace("randomForest", quietly = TRUE)) {
library(randomForest)
# Random Forest comparison
rf.fit <- randomForest(trainX, trainY, ntree = ntrees,
mtry = mtry, nodesize = nmin,
replace = FALSE,
sampsize = trainn*0.8,
importance = TRUE)
par(mfrow=c(1,2))
par(mar = c(1, 2, 2, 2))
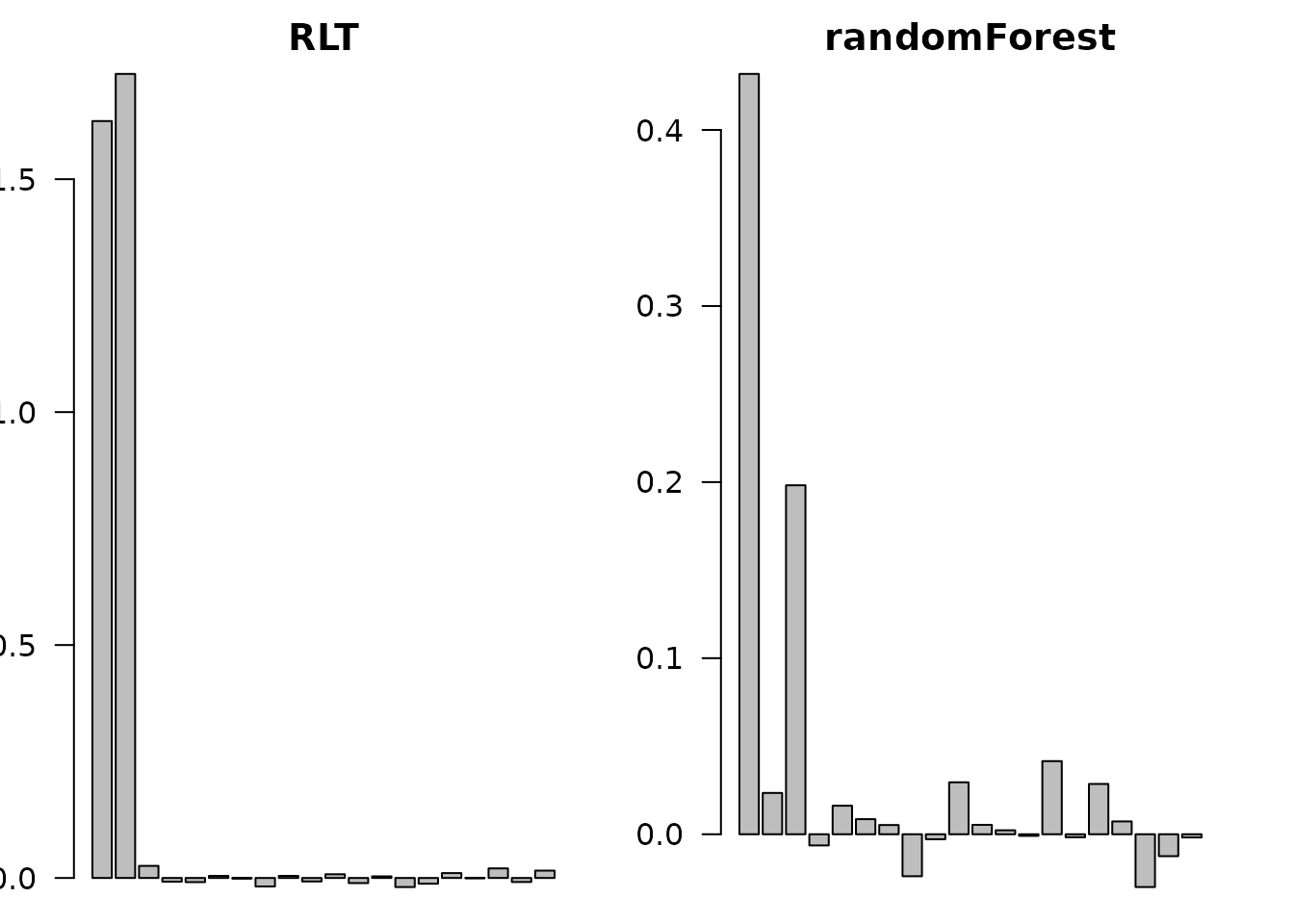
barplot(as.vector(RLTfit$VarImp), main = "RLT",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
barplot(rf.fit$importance[, 1], main = "randomForest",
names.arg = paste0("X", 1:p), las = 2, cex.names = 0.7)
}
Key Points
- Permutation Importance: Requires more out-of-bag samples but provides standard variable importance measures
- Distributed Importance: More efficient for sampling without replacement, can work with fewer samples
- Different Models: Variable importance can be calculated for regression, classification, and survival models
- Splitting Rules: In survival analysis, different splitting rules may produce different variable importance rankings
- Interpretation: Higher importance values indicate variables that have greater influence on predictions