RLT Package Testing Survival Functions and Features
Ruoqing Zhu
Last Updated: July 17, 2025
Test-Surv.RmdInstall and Load Package
Install and load the GitHub version of the RLT package. Do not use the CRAN version.
# install.packages("devtools")
# devtools::install_github("teazrq/RLT")
library(RLT)
## RLT and Random Forests v4.2.6
## pre-release at github.com/teazrq/RLTLoad other packages used in this guide.
library(randomForest)
library(randomForestSRC)
library(ranger)
library(parallel)
library(survival)Benchmark Against Existing Packages
We generate a dataset with 300 observations and 200 variables, with 100 continuous variables and 100 categorical ones with three categories. The survival time and censoring time all follow an exponential distribution.
# generate date
set.seed(1)
trainn = 500
testn = 1000
n = trainn + testn
p = 200
X1 = matrix(rnorm(n*p/2), n, p/2)
X2 = matrix(as.integer(runif(n*p/2)*3), n, p/2)
X = data.frame(X1, X2)
xlink <- function(x) exp(x[, 7] + x[, 16] + x[, 25] + x[, p])
FT = rexp(n, rate = xlink(X))
CT = rexp(n, rate = 0.5)
y = pmin(FT, CT)
Censor = as.numeric(FT <= CT)
mean(Censor)
## [1] 0.744
# parameters
ntrees = 500
ncores = 10
nmin = 25
mtry = p/3
sampleprob = 0.85
rule = "random"
nsplit = ifelse(rule == "best", 0, 3)
importance = TRUE
trainX = X[1:trainn, ]
trainY = y[1:trainn]
trainCensor = Censor[1:trainn]
testX = X[1:testn + trainn, ]
testY = y[1:testn + trainn]
testCensor = Censor[1:testn + trainn]
# get true survival function
timepoints = sort(unique(trainY[trainCensor==1]))
SurvMat = matrix(NA, testn, length(timepoints))
expxlink = xlink(testX)
for (j in 1:length(timepoints))
{
SurvMat[, j] = 1 - pexp(timepoints[j], rate = expxlink )
}
# Calculate timepoint indices for ranger (different from RLT)
yloc = rep(NA, length(timepoints))
for (i in 1:length(timepoints)) yloc[i] = sum( timepoints[i] >= trainY )
for (j in (p/2 + 1):p) X[,j] = as.factor(X[,j])
# recording results
metric = data.frame(matrix(NA, 6, 7))
rownames(metric) = c("rlt", "rltsup", "rltcox", "rltcoxpen", "rsf", "ranger")
colnames(metric) = c("fit.time", "pred.time", "oob.error", "pred.error", "L1",
"obj.size", "tree.size")
# fit RLT with log-rank split
start_time <- Sys.time()
RLTfit.logrank <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, ncores = ncores,
nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = sampleprob,
importance = importance,
param.control = list(split.rule = "logrank", "alpha" = 0.2),
verbose = TRUE, resample.replace=FALSE)
## Fitting Survival Forest...
## ---------- Parameters Summary ----------
## (N, P) = (500, 200)
## # of trees = 500
## (mtry, nmin) = (66, 25)
## split generate = Random, 3
## sampling = 0.85 w/o replace
## (Obs, Var) weights = (No, No)
## alpha = 0.2
## importance = permute
## reinforcement = No
## ----------------------------------------
## Do not have 10 cores, use maximum 4 cores.
metric[1, 1] = difftime(Sys.time(), start_time, units = "secs")
start_time <- Sys.time()
RLTPred <- predict(RLTfit.logrank, testX, ncores = ncores)
metric[1, 2] = difftime(Sys.time(), start_time, units = "secs")
metric[1, 3] = RLTfit.logrank$Error
metric[1, 4] = 1 - cindex(testY, testCensor, colSums(apply(RLTPred$Hazard, 1, cumsum)))
metric[1, 5] = mean(colMeans(abs(RLTPred$Survival - SurvMat)))
metric[1, 6] = format(object.size(RLTfit.logrank), units = "MB")
metric[1, 7] = mean(unlist(lapply(RLTfit.logrank$FittedForest$SplitVar, length)))
# fit RLT with sup-log-rank split
start_time <- Sys.time()
RLTfit.suplogrank <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, ncores = ncores,
nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = sampleprob,
importance = importance,
param.control = list(split.rule = "suplogrank", "alpha" = 0),
verbose = TRUE, resample.replace=FALSE)
## Fitting Survival Forest...
## ---------- Parameters Summary ----------
## (N, P) = (500, 200)
## # of trees = 500
## (mtry, nmin) = (66, 25)
## split generate = Random, 3
## sampling = 0.85 w/o replace
## (Obs, Var) weights = (No, No)
## importance = permute
## reinforcement = No
## ----------------------------------------
## Do not have 10 cores, use maximum 4 cores.
metric[2, 1] = difftime(Sys.time(), start_time, units = "secs")
start_time <- Sys.time()
RLTPred <- predict(RLTfit.suplogrank, testX, ncores = ncores)
metric[2, 2] = difftime(Sys.time(), start_time, units = "secs")
metric[2, 3] = RLTfit.suplogrank$Error
metric[2, 4] = 1- cindex(testY, testCensor, colSums(apply(RLTPred$Hazard, 1, cumsum)))
metric[2, 5] = mean(colMeans(abs(RLTPred$Survival - SurvMat)))
metric[2, 6] = format(object.size(RLTfit.suplogrank), units = "MB")
metric[2, 7] = mean(unlist(lapply(RLTfit.suplogrank$FittedForest$SplitVar, length)))
# fit RLT with cox-grad split
start_time <- Sys.time()
RLTfit.cg <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, ncores = ncores,
nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = sampleprob,
importance = importance,
param.control = list(split.rule = "coxgrad", "alpha" = 0),
verbose = TRUE, resample.replace=FALSE)
## Fitting Survival Forest...
## ---------- Parameters Summary ----------
## (N, P) = (500, 200)
## # of trees = 500
## (mtry, nmin) = (66, 25)
## split generate = Random, 3
## sampling = 0.85 w/o replace
## (Obs, Var) weights = (No, No)
## importance = permute
## reinforcement = No
## ----------------------------------------
## Do not have 10 cores, use maximum 4 cores.
metric[3, 1] = difftime(Sys.time(), start_time, units = "secs")
start_time <- Sys.time()
RLTPred <- predict(RLTfit.cg, testX, ncores = ncores)
metric[3, 2] = difftime(Sys.time(), start_time, units = "secs")
metric[3, 3] = RLTfit.cg$Error
metric[3, 4] = 1 - cindex(testY, testCensor, colSums(apply(RLTPred$Hazard, 1, cumsum)))
metric[3, 5] = mean(colMeans(abs(RLTPred$Survival - SurvMat)))
metric[3, 6] = format(object.size(RLTfit.cg), units = "MB")
metric[3, 7] = mean(unlist(lapply(RLTfit.cg$FittedForest$SplitVar, length)))
# fit RLT with penalized coxgrad split
start_time <- Sys.time()
RLTfit.pcg <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = ntrees, nmin = nmin, mtry = mtry, nsplit = nsplit,
split.gen = rule, resample.prob = sampleprob, importance = importance,
# var.w = ifelse(c(1:(p)) %in% c(7, 16, 25, p), 1, 0.5),
var.w = pmax(max(0, mean(RLTfit.logrank$VarImp)), RLTfit.logrank$VarImp),
param.control = list(split.rule = "coxgrad", "alpha" = 0),
verbose = TRUE, ncores = ncores, resample.replace=FALSE)
## Fitting Survival Forest...
## ---------- Parameters Summary ----------
## (N, P) = (500, 200)
## # of trees = 500
## (mtry, nmin) = (66, 25)
## split generate = Random, 3
## sampling = 0.85 w/o replace
## (Obs, Var) weights = (No, Yes)
## importance = permute
## reinforcement = No
## ----------------------------------------
## Do not have 10 cores, use maximum 4 cores.
metric[4, 1] = difftime(Sys.time(), start_time, units = "secs")
start_time <- Sys.time()
RLTPredp <- predict(RLTfit.pcg, testX, ncores = ncores)
metric[4, 2] = difftime(Sys.time(), start_time, units = "secs")
metric[4, 3] = RLTfit.pcg$Error
metric[4, 4] = 1 - cindex(testY, testCensor, colSums(apply(RLTPredp$Hazard, 1, cumsum)))
metric[4, 5] = mean(colMeans(abs(RLTPredp$Survival - SurvMat)))
metric[4, 6] = format(object.size(RLTfit.pcg), units = "MB")
metric[4, 7] = mean(unlist(lapply(RLTfit.pcg$FittedForest$SplitVar, length)))
# fit rsf
options(rf.cores = ncores)
start_time <- Sys.time()
rsffit <- rfsrc(Surv(trainY, trainCensor) ~ ., data = data.frame(trainX, trainY, trainCensor),
ntree = ntrees, nodesize = nmin, mtry = mtry,
nsplit = nsplit, sampsize = trainn*sampleprob,
importance = ifelse(importance==TRUE,"random", "none"), samptype = "swor",
block.size = 1, ntime = NULL)
metric[5, 1] = difftime(Sys.time(), start_time, units = "secs")
start_time <- Sys.time()
rsfpred = predict(rsffit, data.frame(testX))
metric[5, 2] = difftime(Sys.time(), start_time, units = "secs")
metric[5, 3] = tail(rsffit$err.rate, 1)
metric[5, 4] = 1 - cindex(testY, testCensor, rowSums(rsfpred$chf))
metric[5, 5] = mean(colMeans(abs(rsfpred$survival - SurvMat)))
metric[5, 6] = format(object.size(rsffit), units = "MB")
metric[5, 7] = rsffit$forest$totalNodeCount / rsffit$forest$ntree
# fit ranger
start_time <- Sys.time()
rangerfit <- ranger(Surv(trainY, trainCensor) ~ ., data = data.frame(trainX, trainY, trainCensor),
num.trees = ntrees, min.node.size = nmin, mtry = mtry,
splitrule = "logrank", num.threads = ncores,
sample.fraction = sampleprob, importance = "permutation")
## Growing trees.. Progress: 89%. Estimated remaining time: 3 seconds.
metric[6, 1] = difftime(Sys.time(), start_time, units = "secs")
start_time <- Sys.time()
rangerpred = predict(rangerfit, data.frame(testX))
metric[6, 2] = difftime(Sys.time(), start_time, units = "secs")
metric[6, 3] = rangerfit$prediction.error
metric[6, 4] = 1 - cindex(testY, testCensor, rowSums(rangerpred$chf))
# For ranger, calculate L1 error using available timepoints
# Use the first min(ncol(rangerpred$survival), ncol(SurvMat)) columns
n_cols = min(ncol(rangerpred$survival), ncol(SurvMat))
metric[6, 5] = mean(colMeans(abs(rangerpred$survival[, 1:n_cols] - SurvMat[, 1:n_cols])))
metric[6, 6] = format(object.size(rangerfit), units = "MB")
metric[6, 7] = mean(unlist(lapply(rangerfit$forest$split.varIDs, length)))
metric
## fit.time pred.time oob.error pred.error L1 obj.size tree.size
## rlt 1.9148796 0.3016081 0.2236643 0.2031392 0.1551231 76.4 Mb 85.668
## rltsup 2.4169734 0.3006041 0.2250207 0.2034734 0.1564303 75.4 Mb 84.444
## rltcox 0.9343102 0.2635462 0.2523899 0.2149457 0.1740367 49.4 Mb 54.132
## rltcoxpen 0.8211284 0.2793784 0.2140024 0.1911703 0.1343944 52.4 Mb 57.724
## rsf 11.1473689 0.6099076 0.2276127 0.2088789 0.1497412 68.8 Mb 38.120
## ranger 37.6971779 2.4660072 0.2606490 0.2434724 0.1733328 81.1 Mb 99.680Print a Single Tree
You can use the get.one.tree() function to peek into a
single tree.
# print one tree
tree_num = 1
get.one.tree(RLTfit.logrank, tree_num)
## Tree #1 in the fitted survival forest:
## SplitVar SplitValue LeftNode RightNode NodeWeight
## 1 X43 1.68288969 2 3 0
## 2 X100.1 1.00000000 4 5 0
## 3 <NA> NA NA NA 24
## 4 X41 -0.28036643 6 7 0
## 5 X25 0.55939226 54 55 0
## 6 X7 -0.59370554 8 9 0
## 7 X16 0.69455914 28 29 0
## 8 X13 0.14737996 10 11 0
## 9 X5 -1.72496728 12 13 0
## 10 <NA> NA NA NA 17
## 11 <NA> NA NA NA 14
## 12 <NA> NA NA NA 4
## 13 X58 0.91986866 14 15 0
## 14 X7 0.38743297 16 17 0
## 15 <NA> NA NA NA 14
## 16 X52 1.38145859 18 19 0
## 17 X29 -2.35909845 24 25 0
## 18 X56 -0.07985515 20 21 0
## 19 <NA> NA NA NA 2
## 20 X16 -0.07383513 22 23 0
## 21 <NA> NA NA NA 13
## 22 <NA> NA NA NA 15
## 23 <NA> NA NA NA 12
## 24 <NA> NA NA NA 1
## 25 X25 0.61244108 26 27 0
## 26 <NA> NA NA NA 21
## 27 <NA> NA NA NA 6
## 28 X85 0.52164387 30 31 0
## 29 X21 0.64551395 50 51 0
## 30 X24 -1.64435826 32 33 0
## 31 X19 -0.98728279 46 47 0
## 32 <NA> NA NA NA 3
## 33 X42 0.33495776 34 35 0
## 34 X7 1.17477657 36 37 0
## 35 <NA> NA NA NA 23
## 36 X6 -1.35745218 38 39 0
## 37 <NA> NA NA NA 6
## 38 <NA> NA NA NA 5
## 39 X95 -1.11877220 40 41 0
## 40 <NA> NA NA NA 2
## 41 X91 -1.90238058 42 43 0
## 42 <NA> NA NA NA 2
## 43 X18 -0.21339745 44 45 0
## 44 <NA> NA NA NA 15
## 45 <NA> NA NA NA 17
## 46 <NA> NA NA NA 6
## 47 X42.1 0.00000000 48 49 0
## 48 <NA> NA NA NA 9
## 49 <NA> NA NA NA 23
## 50 X67 1.38454871 52 53 0
## 51 <NA> NA NA NA 8
## 52 <NA> NA NA NA 24
## 53 <NA> NA NA NA 2
## 54 X16 0.52847380 56 57 0
## 55 X33.1 1.00000000 80 81 0
## 56 X81 1.11513339 58 59 0
## 57 X65 0.86251034 74 75 0
## 58 X7 1.98049752 60 61 0
## 59 <NA> NA NA NA 4
## 60 X84 1.26782462 62 63 0
## 61 <NA> NA NA NA 3
## 62 X21 1.39688296 64 65 0
## 63 <NA> NA NA NA 4
## 64 X16 0.10839234 66 67 0
## 65 <NA> NA NA NA 6
## 66 X92 -1.30290477 68 69 0
## 67 <NA> NA NA NA 8
## 68 <NA> NA NA NA 2
## 69 X59 1.47732823 70 71 0
## 70 X47.1 1.00000000 72 73 0
## 71 <NA> NA NA NA 1
## 72 <NA> NA NA NA 23
## 73 <NA> NA NA NA 18
## 74 X55 -1.75655797 76 77 0
## 75 <NA> NA NA NA 1
## 76 <NA> NA NA NA 1
## 77 X7 0.15223985 78 79 0
## 78 <NA> NA NA NA 14
## 79 <NA> NA NA NA 13
## 80 <NA> NA NA NA 23
## 81 <NA> NA NA NA 16
# to get the estimated hazard function of a terminal node
terminal_nodes = RLTfit.logrank$FittedForest$SplitVar[[tree_num]] == -1
haz = RLTfit.logrank$FittedForest$NodeHaz[[tree_num]][[which(terminal_nodes)[1]]]
# to get the estimated survival function of a terminal node
plot(c(0, RLTfit.logrank$timepoints),
exp(-cumsum(haz)),
xlab = "time", ylab = "survival", type = "l",
main = paste("First terminal node of Tree", tree_num))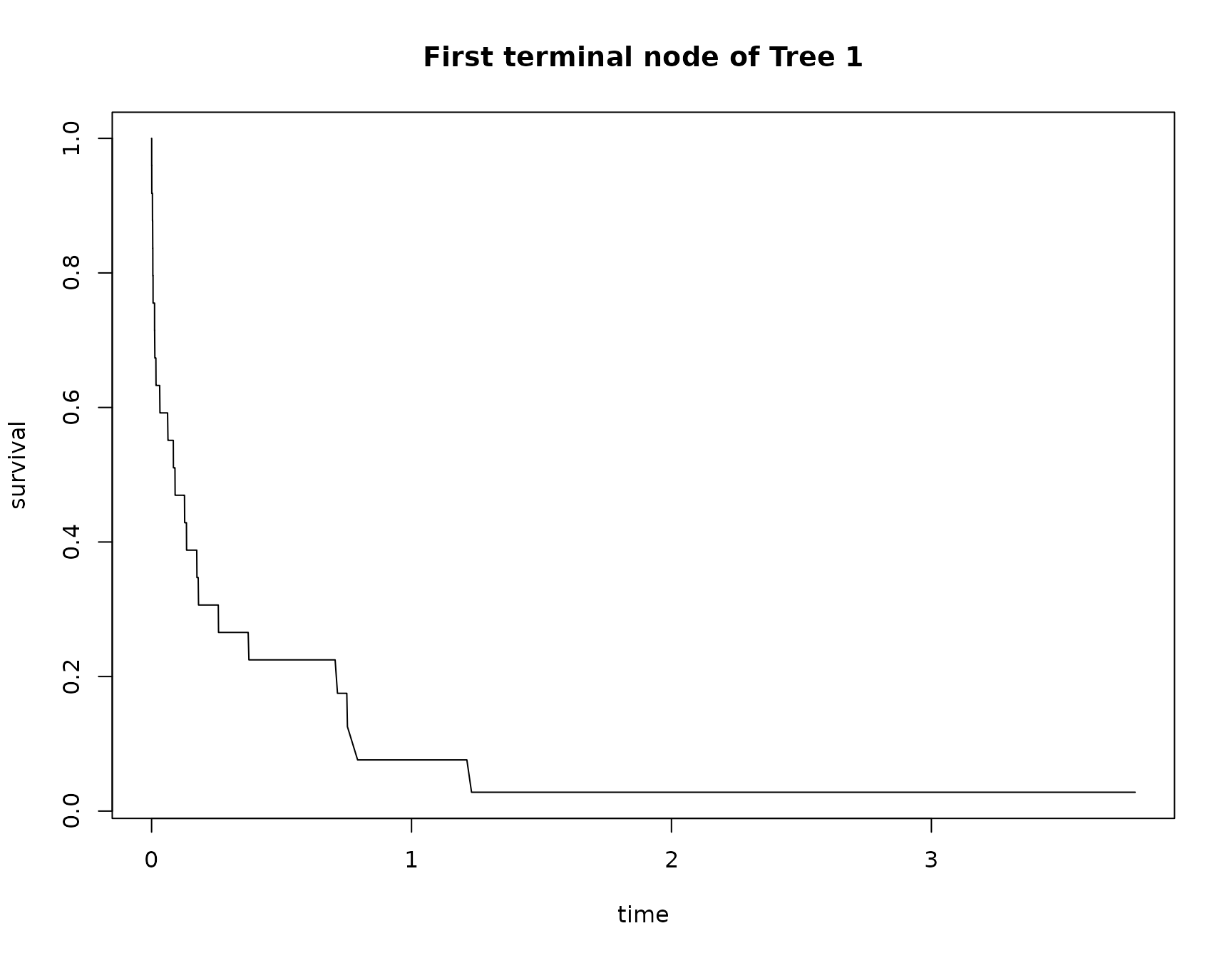
Random Forest Kernel
KernelW = forest.kernel(RLTfit.logrank, X1 = testX[1, ], X2 = trainX)$KernelSetting random seed
## Fitting a forest
RLTfit1 <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = 100, importance = TRUE, nmin = 1)
RLTfit2 <- RLT(trainX, trainY, trainCensor, model = "survival",
ntrees = 100, importance = TRUE, nmin = 1,
seed = RLTfit1$parameters$seed)
# check if importance are identical
all(RLTfit1$VarImp == RLTfit2$VarImp)
## [1] TRUE
# prediction
RLTPred1 <- predict(RLTfit1, testX, keep.all = TRUE)
RLTPred2 <- predict(RLTfit2, testX, keep.all = TRUE)
# check predictions are identical
all(RLTPred1$Prediction == RLTPred2$Prediction)
## [1] TRUE